paired end sequencing wikipedia
Read length is a factor which can affect the results of biological studies. Paired end mate pair sequencing explanation biocc paired end or mate pair refers to how the library is made and then how it is sequenced.
Output per run for single-end sequencing are noted.

. Briefly the target genomic DNA is isolated and partially digested with restrictio. Paired-end sequencing involves sequencing both ends of a fragment and facilitates detection of genomic rearrangements as well as gene fusions and novel transcripts. Alba by single-molecule real-time technology.
For the first test I took some sequence from the human genome hg19 and created two 100 bp reads from this region. In this study we sequenced the genome of P. The paired-end module allows for up to 36 bases of each end to be sequenced during the same sequencing run.
Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of. However in many cases eg with Illumina NextSeq and NovaSeq. 4516 of the genome was annotated as repetitive.
Specifically the invention provides cloning and DNA manipulation strategies to isolate the two ends of a large target nucleic acid into a single small DNA construct for rapid cloning. For your De novo genome assembly Fig. De novo assembly of P.
Paired end sequencing wikipedia Tuesday June 7 2022 Edit. Both are methodologies that in addition to the sequence information give you information about the physical distance between the two reads in your genome. Now lets get started.
Below is a list of paired end sequencing words - that is words related to paired end sequencing. This gives more specificity to alignment algorithms especially in highly repetitive regions. Any platform that can allow for the ligated fragments to be sequenced across the NheI junction Roche 454 or by paired-end or mate-paired reads Illumina GA and HiSeq platforms would be suitable for Hi-C.
The paired-end module of the GAII system allows both ends of a DNA fragment to be sequenced. All Illumina next-generation sequencing NGS systems are capable of paired-end. The structure of a paired-end read is described here.
Sanger paired ends are generated from a completely different process sequencing the ends of BAC clones and the result is that those paired ends are outie This leads to a lot of confusion when using a mix of technologies or using software that expects your paired ends in a certain orientation. The present invention provides for a method of preparing a target nucleic acid fragments to produce a smaller nucleic acid which comprises the two ends of the target nucleic acid. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts.
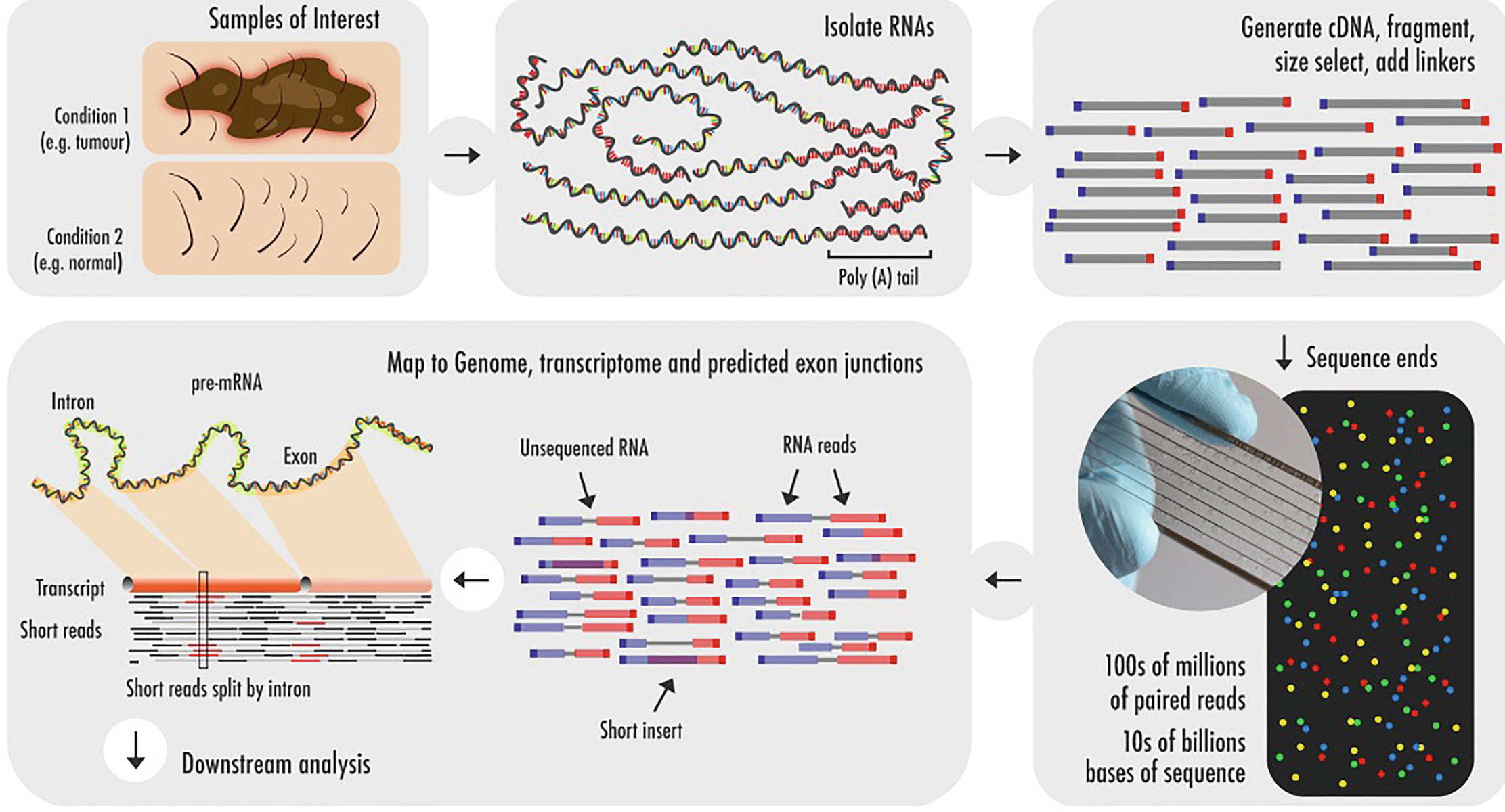
Alba had a genome size of 41599 Mb with a contig N50 of 118 Mb. Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms. There are 38 paired end sequencing-related words in total with the top 5 most semantically related being paired-end tags sequence assembly genetics sequencing and dnaYou can get the definitions of a word in the list below by tapping the question-mark icon.
End-sequence profiling ESP sometimes Paired-end mapping PEM is a method based on sequence-tagged connectors developed to facilitate de novo genome sequencing to identify high-resolution copy number and structural aberrations such as inversions and translocations. Furthermore sequencing can involve single-end SE or paired-end PE readsPaired-end sequencing means sequencing both ends of the cDNA fragments and aligning the forward and. Run times and outputs approximately double when performing paired-end sequencing.
In short-read sequencing intact genomic DNA is sheared into several million short DNA fragments called reads. For mRNA-Seq library prep use. Another consideration is whether to generate a strand-specific library that retains the orientation of the original RNA transcript which may be critical to identify antisense or non-coding RNA.
Longer runs up to 72nt should be available in. Since paired-end reads are more likely to align to a reference the quality of the entire data set improves. For sequencing projects that require higher accuracy such as studies of alternate splicing 40 million to 60 million paired-end reads will provide better results.
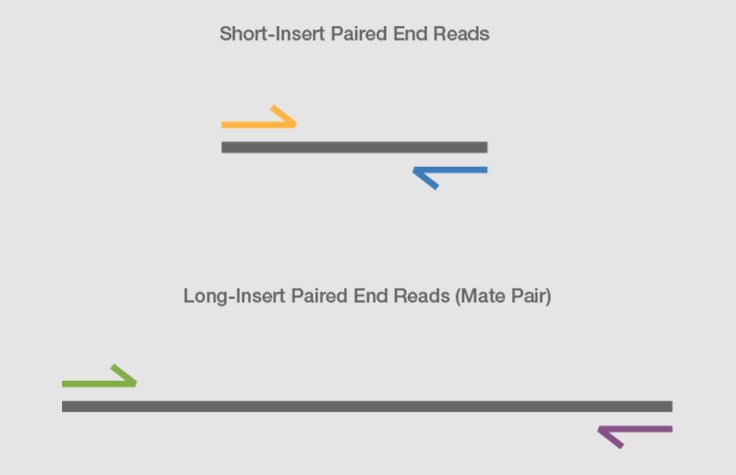
For more detailed analyses to determine for example allele-specific expression or expression of low-abundant transcripts 60 million to 100 million reads may be required. Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased information for maximising sequencing coverage across a genome 1. It is named by analogy with the rapidly expanding quasi-random shot grouping of a shotgun.
Average read lengths for the Roche 454 and Helicos. This can be very helpful e. Before high-throughput sequencing the quality of the library should be verified using Sanger sequencing wherein the long sequencing read.
The inner mate distance between the two reads is 200 bp creating an insert size of 400 bp. The chain-termination method of DNA sequencing Sanger sequencing can only be used for short DNA strands of 100 to 1000 base pairsDue to this size limit longer sequences are subdivided. In genetics shotgun sequencing is a method used for sequencing random DNA strands.
Individual reads can be paired together to create paired-end reads which offers some benefits for downstream bioinformatics data analysis algorithms. Mate pair sequencing is used for various applications applications including. It also can be cultivated and grown well in northern China.
A total of 32963 protein-coding genes were identified. Sequencing technologies vary in the length of reads produced. The reads were then mapped back to the reference using BWA aln and sampe.
Typical experimental design advice for expression analyses using RNA-seq generally assumes that single-end reads provide robust gene-level expression estimates in a cost-effective manner and that the additional benefits obtained from paired-end sequencing are not worth the additional cost. For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing.

Paired End Shotgun Sequencing Okinawa Institute Of Science And Technology Graduate University Oist
Difference Between Whole Genome Sequencing And Shotgun Sequencing Difference Between
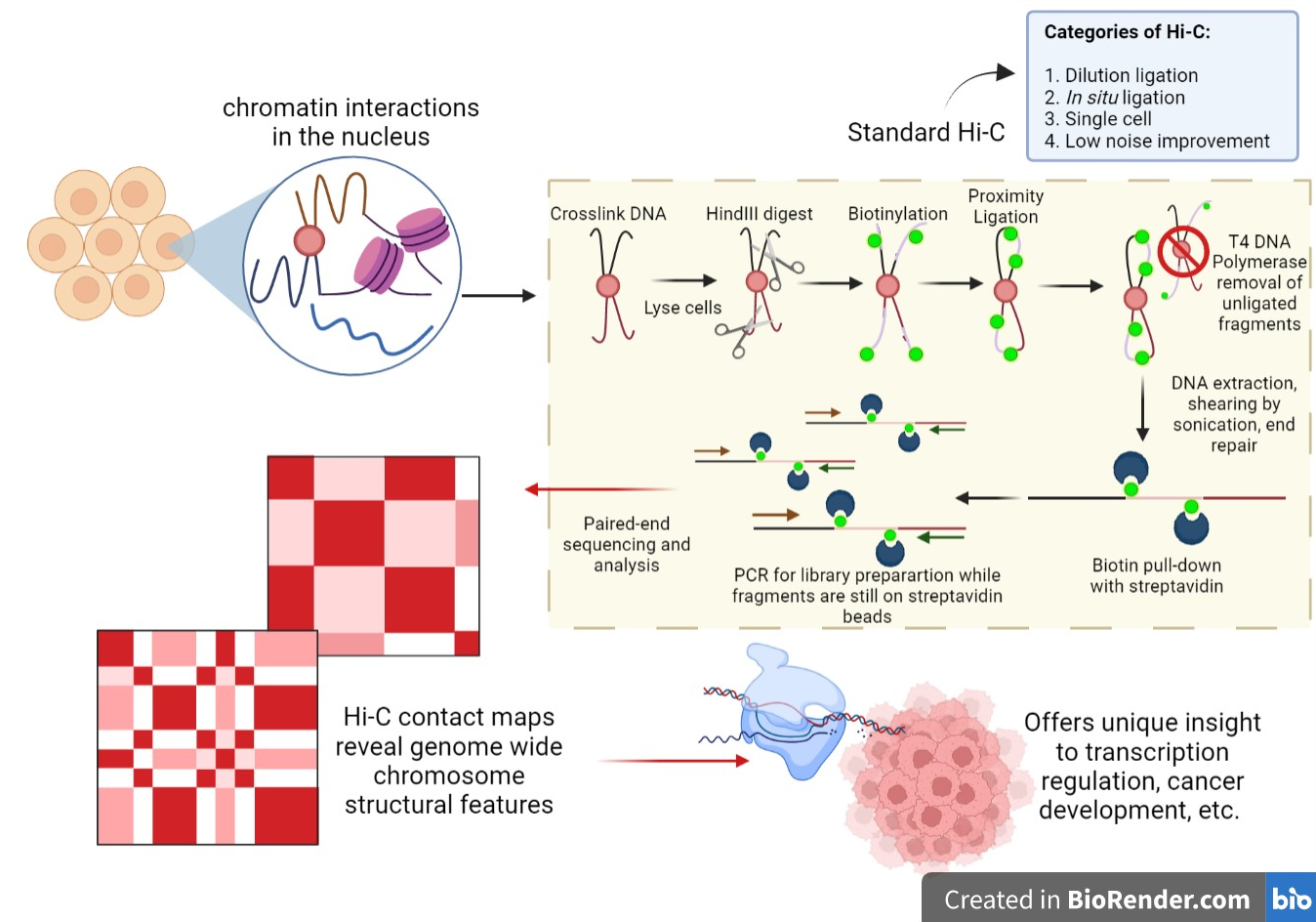
Hi C Genomic Analysis Technique Wikipedia

Bacterial Chromosome Replication Biology Lessons Chromosome Biology

Next Generation Sequencing And Its Applications Sciencedirect

What Is Mate Pair Sequencing For

What Are Paired End Reads The Sequencing Center

What Are Paired End Reads The Sequencing Center

What Is Mate Pair Sequencing For

Bacterial Chromosome Replication Biology Lessons Chromosome Biology

Sequencing Introduction To Sequencing By Synthesis Sbs Youtube
Sequencing Introduction To Sequencing By Synthesis Sbs Youtube